Build your own expression explorer with OmicsEE
OmicsEE is a browser-based application designed to facilitate the interactive exploration of omics expression data across multiple atlases.
Read more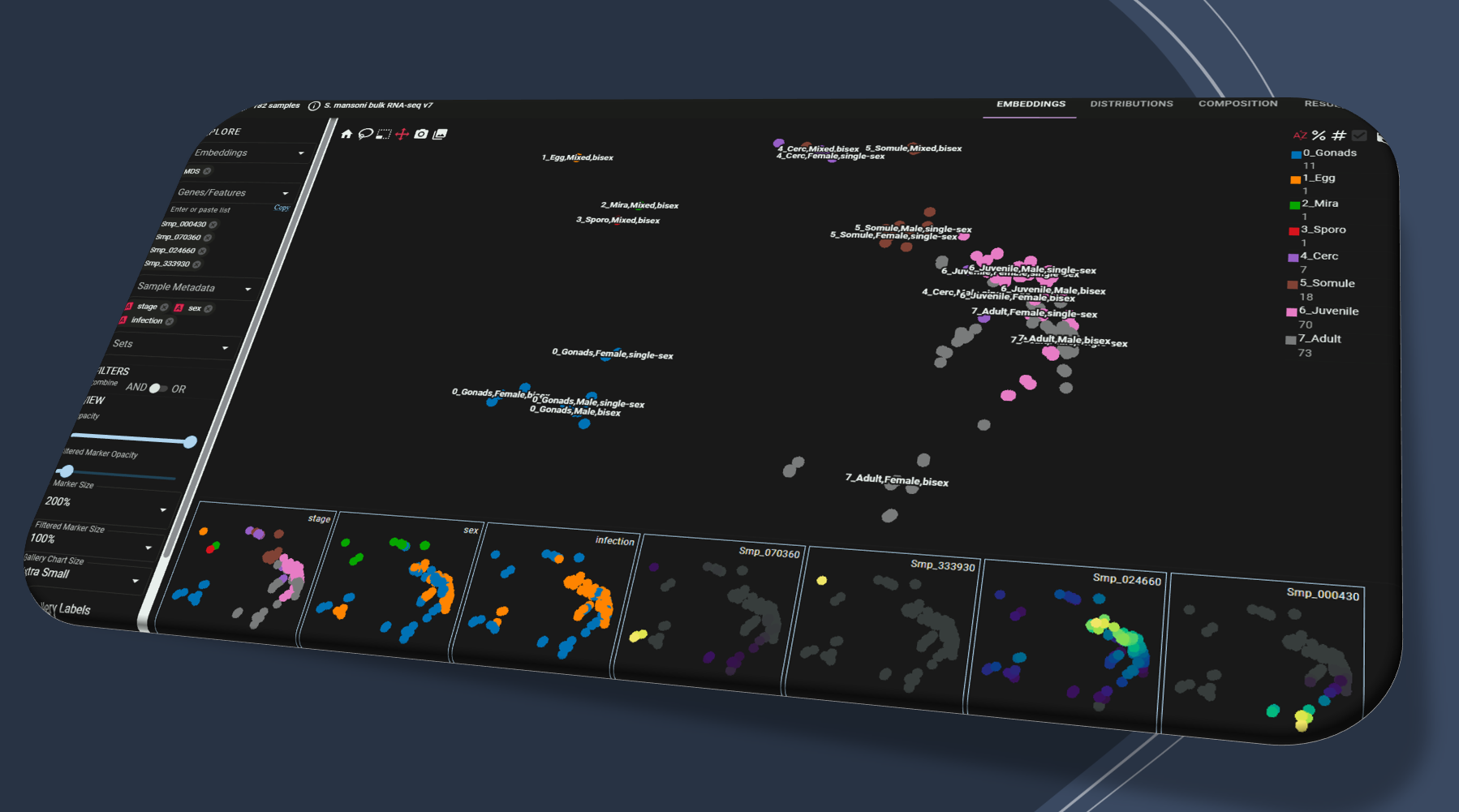
public and re-analyzed; multi-species; at the levels of bulk, single-cell, and spatial atlases!
Explore Schisto.xyz V2(Multiple) Gene queries, flexible embeddings and metadata.
Share the current visualization state easily in a URL.
Fast expression exploration in multiple datasets simultaneously.
Serving schisto researchers since 2018, now offers enhanced datasets, interactive visualisation solutions, and a re-designed powerful search engine.
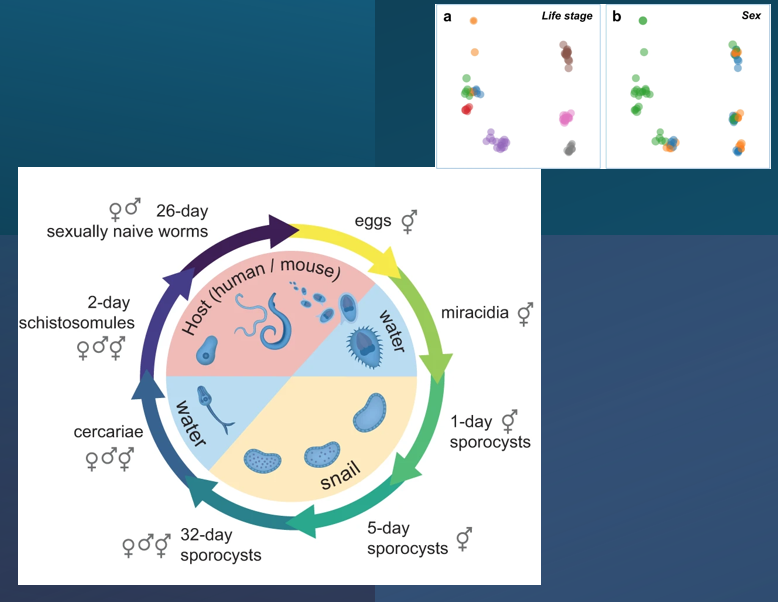
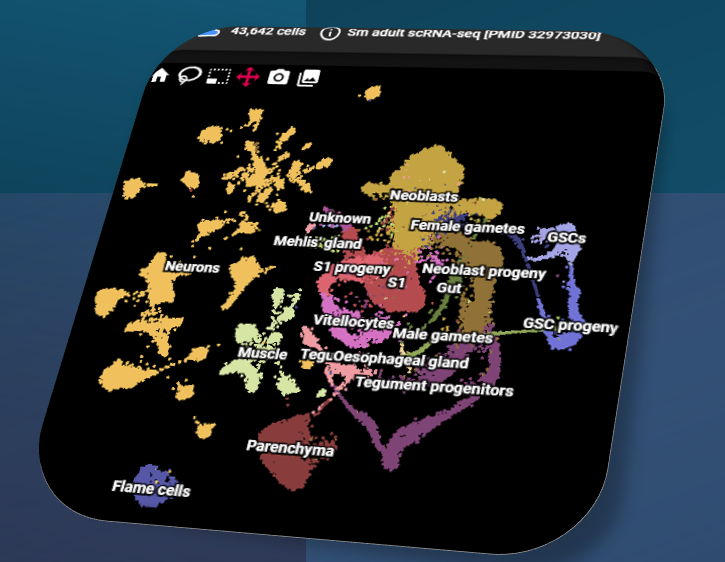
Bulk RNA-seq for all life stages; integrative re-analysis; single-cell atlases; spatial expression (ITF).
Browser-based interactive visualisation supports multiple queries and embeddings, easy share of states via URLs.
Metadata selection, point colors/sizes, subsetting, distribution and composition plots, all highly customizable.
The search engine has been re-designed to provide an expression overview across multiple datasets, fast and robust.
Fast visualisation of expression in millions of cells / samples, across multiple datasets, at one time.
Explore your favourite gene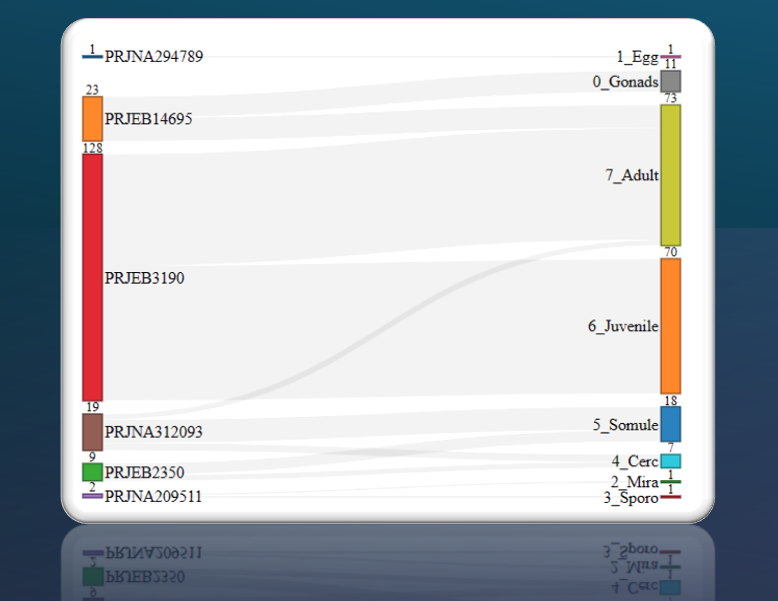



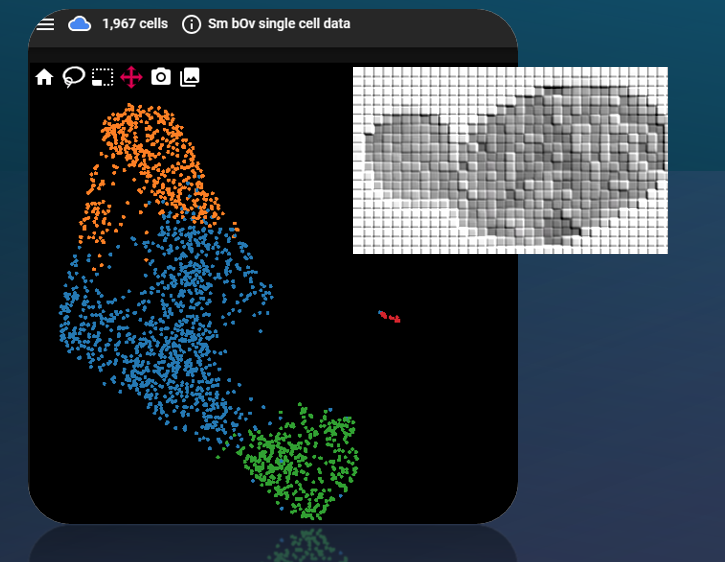
Sj bulk RNA-seq data from Cheng et al 2022 PMID:35025881, 12 larval samples.
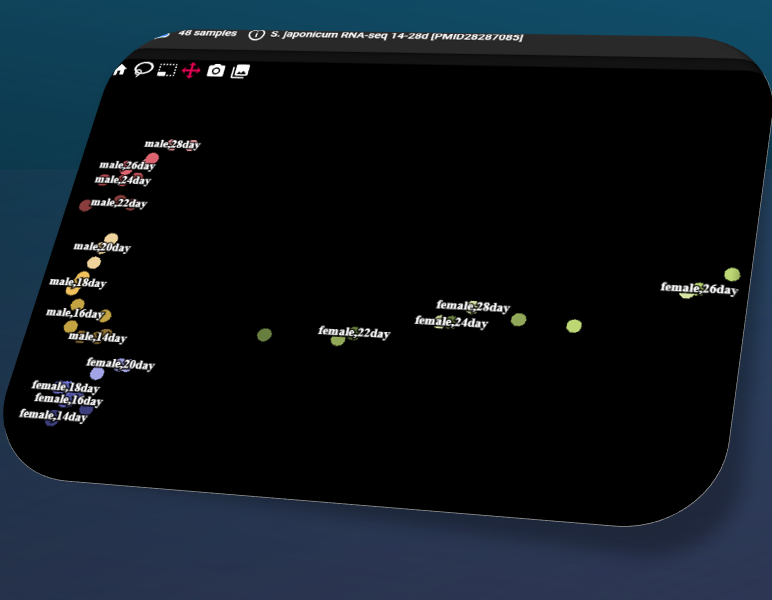
Sj bulk RNA-seq data from Wang et al 2017 PMID:28287085, 48 samples 14-28dpi, re-processed

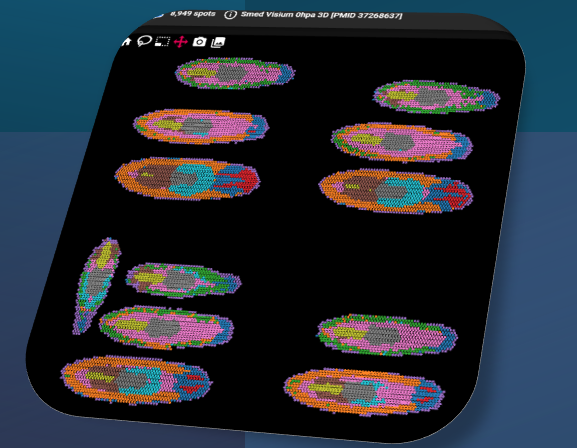
Planarian 55K cells at different regeneration time, norm counts. PMID:37268637
The retinoic acid family-like nuclear receptor SmRAR identified by single-cell transcriptomics of ovarian cells controls oocyte differentiation in Schistosoma mansoni
A novel, non-neuronal acetylcholinesterase of schistosome parasites is essential for definitive host infection
Convergent evolution of a genotoxic stress response in a parasite-specific p53 homolog
Rhesus macaques self-curing from a schistosome infection can display complete immunity to challenge
Schistosoma mansoni α-N-acetylgalactosaminidase (SmNAGAL) regulates coordinated parasite movement and egg production
Programmed genome editing of the omega-1 ribonuclease of the blood fluke, Schistosoma mansoni
OmicsEE is a browser-based application designed to facilitate the interactive exploration of omics expression data across multiple atlases.
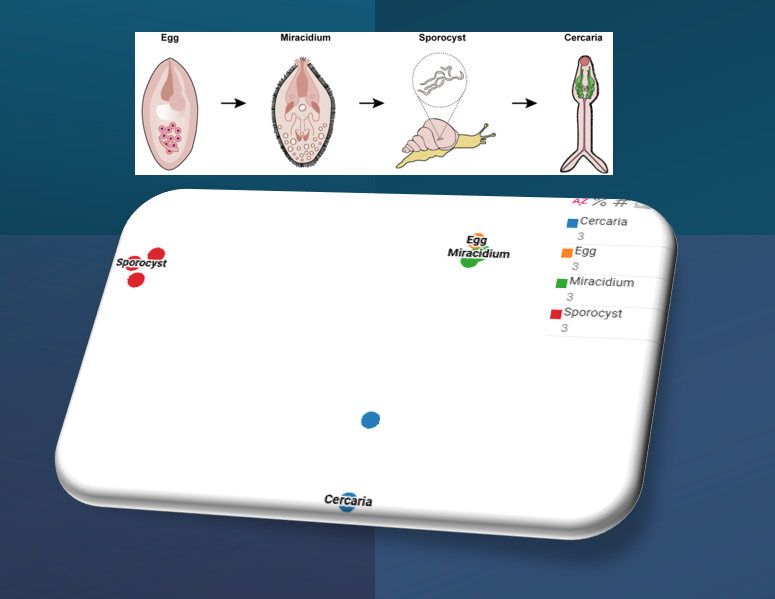
Read moreSingle-cell datasets for S. mansoni miracidia (20K cells) and D5 sporocysts (601 cells) are available now!
Read moreS. japonicum bulk RNA-seq covering larval stages from Cheng et al 2022. Data was mapped to the HuSjv3 genome and TPM was used for normalization.
Read moreFeel free to drop me a message via social media / email / contact form. If you find the resource useful, please cite (for now)







SchistoXYZ, as a valuable resource for the research community, has contributed to more than 30 published studies. Here are a few selections