The study provided multiple Visium sections for Schmidtea mediterranea at 0hpa. On the scanpy data I modified the spatial coordinates for a better view, and also constructed their 3D structure.
Here is the link to the original publication in Nat Commu: https://www.nature.com/articles/s41467-023-39016-0
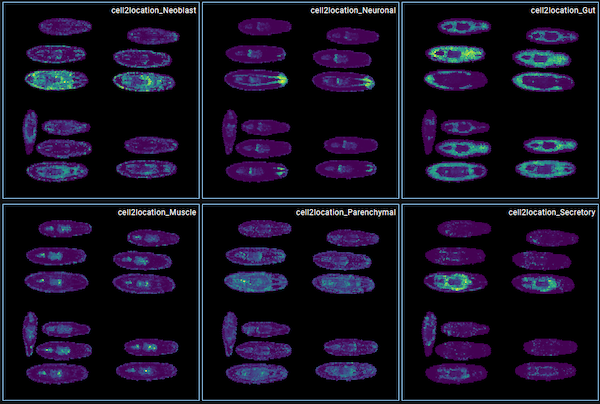
They integrated all sections from the same timepoint, eg. 12 sections at 0 h post amputation (hpa), used STAGATE to identify spatial domains including Body wall, Gut, CNS, Pharynx etc, and used their single-cell data to deconvolute the visium spots using Cell2Location. Corresponding H&E images were also accessible to identify the tissue structure.

The authors also offered a resource to visualise the data (https://ngdc.cncb.ac.cn/STAPR/) but unfortunately the plotting functions seem not working. Therefore I tried to re-construct the 3D structure based on the indicated coordinates, it works in principle but since there are only 12 sections it looks not very vertical.

The authors mapped their omics data to the smed_20140614 genome and annotation (SMED300XXXXX), which seems pretty old and I haven’t found a straightforward way to identify orthologues to S. mansoni, otherwise it would be much easier to check spatial gene expression in this free-living relative.
update 20240911
added the mapping ids from the dd_Smed_v4/v6 transcriptome, as indicated in the smed_20140614.mapping.rosettastone.2020.txt.