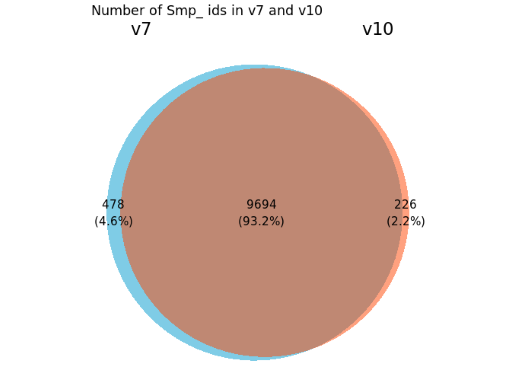
While there is a hugo improvement in the genome assembly from V7 to V10, the geneset stays largely the same.
In the current expression database there are data processed with v7 (eg. integrative bulk RNA-seq, somule scRNA-seq, and adult scRNA-seq) and some with the latest v10 (eg. lifecycle bulk RNA-seq, miracidium and sporocyst scRNA-seq).
I checked the Smp_ identifiers between these two versions: in v7 (v7.2; WBPS14) there are 10,172 genes and in v10 (WBPS19) 9,920 genes. About 97.7% Smp ids exist in both annotations (normally the gene model doesn’t change much if the Smp id stays the same) and 226 Smp_ ids were unique in v10 (2.3%), which can be new or curated gene models.
> table(v10only$chr)
SM_V10_1 SM_V10_2 SM_V10_3 SM_V10_4 SM_V10_5 SM_V10_6 SM_V10_7 SM_V10_WSR SM_V10_Z
37 18 28 24 35 13 11 14 46